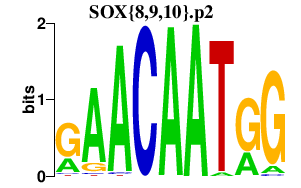
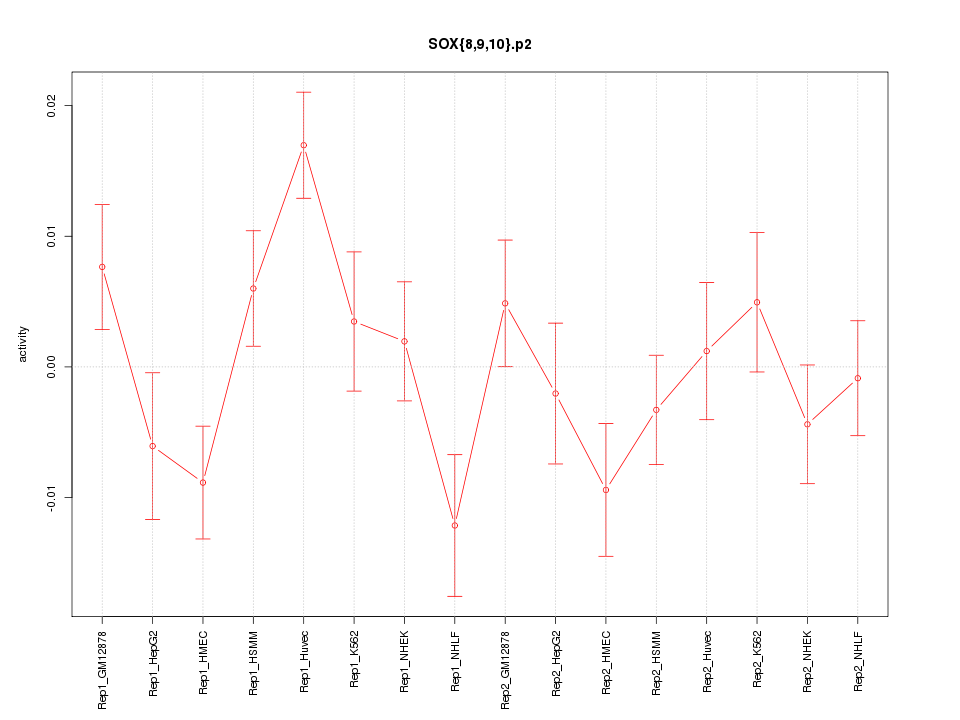
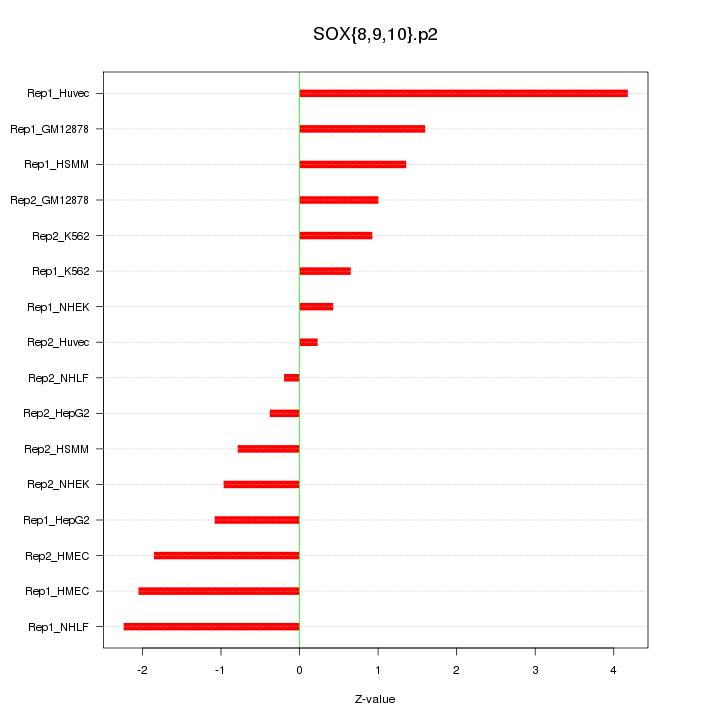
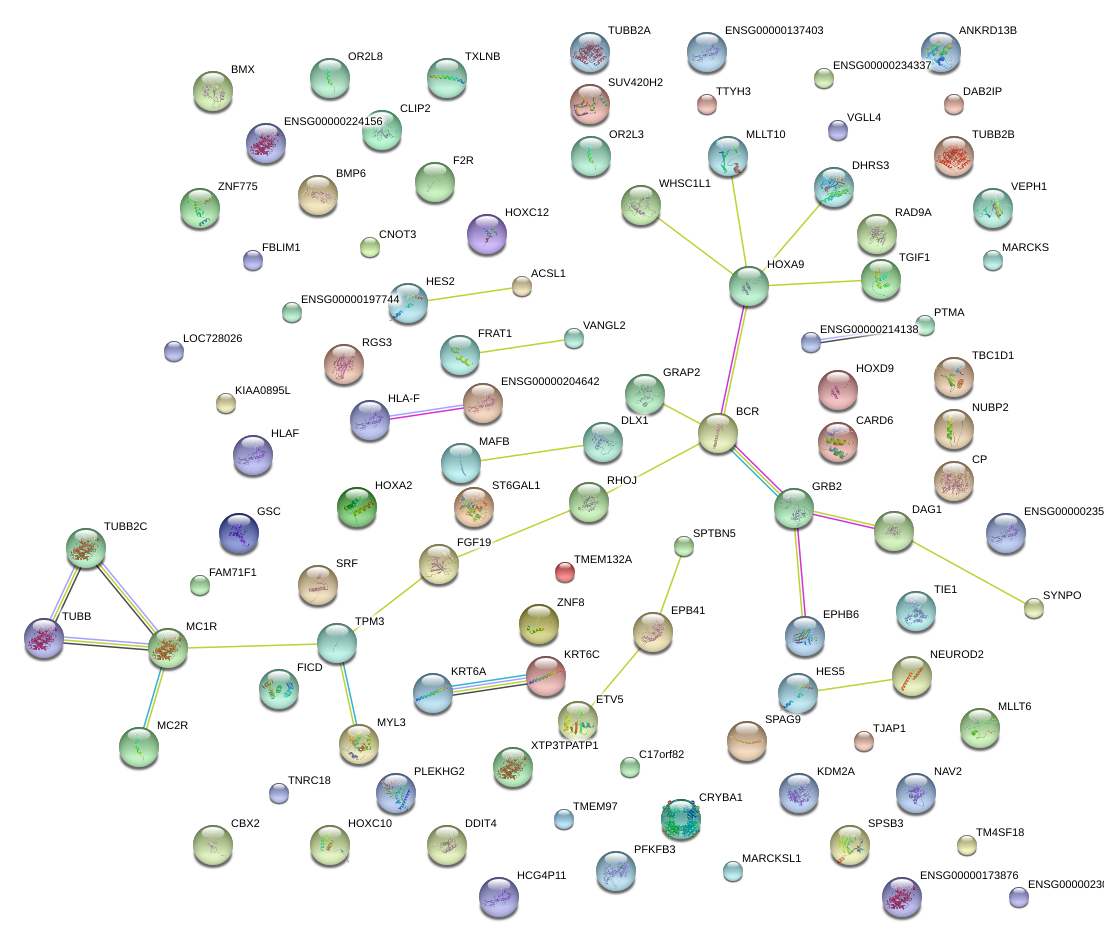
|
chr12_+_52665196
|
1.828
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr9_+_123501186
|
1.725
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr7_+_2638156
|
1.661
|
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr2_+_232281491
|
1.639
|
|
PTMA
|
prothymosin, alpha
|
|
chr20_-_38751289
|
1.587
|
NM_005461
|
MAFB
|
v-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian)
|
|
chr3_-_11598829
|
1.583
|
NM_001128220
|
VGLL4
|
vestigial like 4 (Drosophila)
|
|
chr12_-_120725210
|
1.494
|
|
LOC338799
|
hypothetical LOC338799
|
|
chr3_-_187309442
|
1.481
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr2_+_232281470
|
1.420
|
NM_001099285
NM_002823
|
PTMA
|
prothymosin, alpha
|
|
chr1_-_32574185
|
1.363
|
NM_023009
|
MARCKSL1
|
MARCKS-like 1
|
|
chr11_+_20005704
|
1.355
|
|
NAV2
|
neuron navigator 2
|
|
chr9_+_115303527
|
1.314
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr1_+_43539239
|
1.278
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr7_-_27171673
|
1.248
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr17_+_75366524
|
1.232
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr7_+_2638108
|
1.213
|
NM_025250
|
TTYH3
|
tweety homolog 3 (Drosophila)
|
|
chr1_-_2451543
|
1.200
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr6_+_30796064
|
1.175
|
|
TUBB
|
tubulin, beta
|
|
chr1_+_158636987
|
1.171
|
NM_020335
|
VANGL2
|
vang-like 2 (van gogh, Drosophila)
|
|
chr6_+_43565192
|
1.149
|
NM_001146017
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr1_-_152431175
|
1.118
|
NM_152263
|
TPM3
|
tropomyosin 3
|
|
chr1_+_32992131
|
1.110
|
|
|
|
|
chr14_+_62740878
|
1.078
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr5_+_40877053
|
1.076
|
NM_032587
|
CARD6
|
caspase recruitment domain family, member 6
|
|
chr7_-_27171648
|
1.069
|
|
HOXA9
|
homeobox A9
|
|
chr6_+_114285209
|
1.038
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr18_-_13905534
|
1.033
|
NM_000529
|
MC2R
|
melanocortin 2 receptor (adrenocorticotropic hormone)
|
|
chr17_-_46479127
|
1.028
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr19_+_59333287
|
1.020
|
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr17_+_34115383
|
1.008
|
NM_005937
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr16_-_1772305
|
1.005
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr7_-_27171600
|
0.995
|
|
HOXA9
|
homeobox A9
|
|
chr19_+_59333232
|
0.967
|
NM_014516
|
CNOT3
|
CCR4-NOT transcription complex, subunit 3
|
|
chr7_-_5429702
|
0.952
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr4_-_185963859
|
0.937
|
|
ACSL1
|
acyl-CoA synthetase long-chain family member 1
|
|
chr7_+_148613304
|
0.935
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr10_+_6284845
|
0.933
|
NM_004566
|
PFKFB3
|
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3
|
|
chr3_-_46879883
|
0.928
|
NM_000258
|
MYL3
|
myosin, light chain 3, alkali; ventricular, skeletal, slow
|
|
chr22_+_38652555
|
0.921
|
|
GRAP2
|
GRB2-related adaptor protein 2
|
|
chr1_+_15957832
|
0.920
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr1_+_29113589
|
0.908
|
NM_001166006
|
EPB41
|
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked)
|
|
chr7_+_142270634
|
0.884
|
|
EPHB6
|
EPH receptor B6
|
|
chr16_-_30841938
|
0.884
|
|
NCRNA00095
|
non-protein coding RNA 95
|
|
chr10_+_73703682
|
0.880
|
NM_019058
|
DDIT4
|
DNA-damage-inducible transcript 4
|
|
chr11_+_66764060
|
0.876
|
|
LOC100131150
|
hypothetical protein LOC100131150
|
|
chr7_+_73341640
|
0.864
|
NM_003388
NM_032421
|
CLIP2
|
CAP-GLY domain containing linker protein 2
|
|
chr17_+_56843893
|
0.858
|
NM_203425
|
C17orf82
|
chromosome 17 open reading frame 82
|
|
chr3_-_158704056
|
0.848
|
NM_001167915
NM_001167916
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr11_+_66764064
|
0.833
|
|
KDM2A
|
lysine (K)-specific demethylase 2A
|
|
chr6_+_7671267
|
0.832
|
|
BMP6
|
bone morphogenetic protein 6
|
|
chr6_-_139654807
|
0.821
|
NM_153235
|
TXLNB
|
taxilin beta
|
|
chr19_+_44595589
|
0.801
|
NM_022835
|
PLEKHG2
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 2
|
|
chr12_+_107433179
|
0.798
|
NM_007076
|
FICD
|
FIC domain containing
|
|
chr1_+_246178701
|
0.794
|
NM_001001963
|
OR2L8
|
olfactory receptor, family 2, subfamily L, member 8
|
|
chr7_+_128136347
|
0.775
|
|
FAM71F1
|
family with sequence similarity 71, member F1
|
|
chr4_+_37655385
|
0.774
|
|
TBC1D1
|
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1
|
|
chr6_+_30796123
|
0.772
|
NM_178014
|
TUBB
|
tubulin, beta
|
|
chr3_+_188131209
|
0.769
|
|
ST6GAL1
|
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1
|
|
chr1_-_12599934
|
0.768
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr7_+_149707338
|
0.755
|
NM_173680
|
ZNF775
|
zinc finger protein 775
|
|
chr6_+_29799095
|
0.746
|
NM_001098478
NM_001098479
NM_018950
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr19_+_60543032
|
0.745
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr1_-_6402467
|
0.743
|
NM_019089
|
HES2
|
hairy and enhancer of split 2 (Drosophila)
|
|
chr19_+_63482118
|
0.742
|
NM_021089
|
ZNF8
|
zinc finger protein 8
|
|
chr15_-_39973566
|
0.740
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr8_-_38324809
|
0.727
|
|
WHSC1L1
|
Wolf-Hirschhorn syndrome candidate 1-like 1
|
|
chr16_+_1772929
|
0.724
|
NM_012225
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr6_+_29799182
|
0.720
|
|
HLA-F
|
major histocompatibility complex, class I, F
|
|
chr3_+_49482474
|
0.716
|
NM_001165928
NM_001177634
NM_001177635
NM_001177636
NM_001177637
NM_001177638
NM_001177639
NM_001177640
NM_001177641
NM_001177642
NM_001177644
NM_004393
|
DAG1
|
dystroglycan 1 (dystrophin-associated glycoprotein 1)
|
|
chr5_+_76047536
|
0.711
|
NM_001992
|
F2R
|
coagulation factor II (thrombin) receptor
|
|
chr16_-_65774904
|
0.706
|
|
KIAA0895L
|
KIAA0895-like
|
|
chr3_-_150422474
|
0.706
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chrX_+_15428820
|
0.703
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr17_+_23670247
|
0.694
|
NM_014573
|
TMEM97
|
transmembrane protein 97
|
|
chr1_+_39647762
|
0.694
|
NM_015038
|
KIAA0754
|
KIAA0754
|
|
chr6_+_30796121
|
0.684
|
|
TUBB
|
tubulin, beta
|
|
chr5_+_149960820
|
0.684
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chr3_-_158703821
|
0.681
|
NM_001167911
NM_001167912
|
VEPH1
|
ventricular zone expressed PH domain homolog 1 (zebrafish)
|
|
chr17_+_24598000
|
0.680
|
NM_005208
|
CRYBA1
|
crystallin, beta A1
|
|
chr17_-_70901303
|
0.672
|
|
GRB2
|
growth factor receptor-bound protein 2
|
|
chr11_+_66915997
|
0.670
|
NM_004584
|
RAD9A
|
RAD9 homolog A (S. pombe)
|
|
chr22_+_21852911
|
0.669
|
|
BCR
|
breakpoint cluster region
|
|
chr18_+_3437607
|
0.664
|
NM_173207
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr12_-_51173238
|
0.664
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr17_-_35017691
|
0.663
|
NM_006160
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr7_-_96489884
|
0.662
|
|
|
|
|
chr14_-_94306104
|
0.662
|
NM_173849
|
GSC
|
goosecoid homeobox
|
|
chr17_+_24944605
|
0.656
|
NM_152345
|
ANKRD13B
|
ankyrin repeat domain 13B
|
|
chr10_+_99069007
|
0.653
|
NM_005479
|
FRAT1
|
frequently rearranged in advanced T-cell lymphomas
|
|
chr17_-_35017650
|
0.647
|
|
NEUROD2
|
neurogenic differentiation 2
|
|
chr17_+_37373195
|
0.640
|
|
|
|
|
chr3_-_150533948
|
0.638
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr11_+_60448487
|
0.637
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr11_-_69228286
|
0.635
|
NM_005117
|
FGF19
|
fibroblast growth factor 19
|
|
chr7_-_27108826
|
0.624
|
NM_006735
|
HOXA2
|
homeobox A2
|
|
chr2_+_172658453
|
0.623
|
NM_001038493
NM_178120
|
DLX1
|
distal-less homeobox 1
|
|
chr12_+_52634950
|
0.622
|
NM_173860
|
HOXC12
|
homeobox C12
|
|
chr6_+_43247179
|
0.617
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr16_-_1772577
|
0.617
|
NM_080861
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr1_+_153846602
|
0.616
|
NM_018116
|
MSTO1
|
misato homolog 1 (Drosophila)
|
|
chr21_-_31853160
|
0.613
|
NM_003253
|
TIAM1
|
T-cell lymphoma invasion and metastasis 1
|
|
chr15_+_82945223
|
0.608
|
NM_001007072
NM_017894
NM_181877
|
ZSCAN2
|
zinc finger and SCAN domain containing 2
|
|
chr6_-_10523424
|
0.602
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr22_-_41672990
|
0.602
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr11_+_71524403
|
0.601
|
NM_000804
|
FOLR3
|
folate receptor 3 (gamma)
|
|
chr17_-_1195096
|
0.599
|
|
PAFAH1B1
|
platelet-activating factor acetylhydrolase 1b, regulatory subunit 1 (45kDa)
|
|
chr19_+_53664276
|
0.596
|
NM_004228
NM_017457
|
CYTH2
|
cytohesin 2
|
|
chr6_+_43247168
|
0.595
|
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr1_-_16033866
|
0.595
|
|
|
|
|
chr11_-_27450808
|
0.594
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr17_-_1359745
|
0.592
|
|
INPP5K
|
inositol polyphosphate-5-phosphatase K
|
|
chr8_-_22044463
|
0.589
|
NM_005144
NM_018411
|
HR
|
hairless homolog (mouse)
|
|
chr6_+_43246897
|
0.588
|
NM_003131
|
SRF
|
serum response factor (c-fos serum response element-binding transcription factor)
|
|
chr12_-_69289889
|
0.581
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr14_+_58725104
|
0.578
|
NM_014992
|
DAAM1
|
dishevelled associated activator of morphogenesis 1
|
|
chr12_+_47498778
|
0.575
|
NM_000725
|
CACNB3
|
calcium channel, voltage-dependent, beta 3 subunit
|
|
chr1_-_50661716
|
0.574
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr9_+_133259330
|
0.572
|
|
PRRC2B
|
proline-rich coiled-coil 2B
|
|
chr3_-_50340667
|
0.571
|
NM_007275
|
TUSC2
|
tumor suppressor candidate 2
|
|
chr17_-_37196463
|
0.565
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr19_-_59476734
|
0.564
|
NM_001080978
NM_005874
|
LILRB2
|
leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2
|
|
chr17_+_78009819
|
0.564
|
NM_001083608
NM_012336
NM_031968
|
NARF
|
nuclear prelamin A recognition factor
|
|
chr16_-_1772572
|
0.562
|
|
SPSB3
|
splA/ryanodine receptor domain and SOCS box containing 3
|
|
chr8_-_81155564
|
0.560
|
NM_001025252
|
TPD52
|
tumor protein D52
|
|
chr16_+_464844
|
0.559
|
NM_001142272
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr11_-_16380967
|
0.558
|
NM_001145819
NM_017508
|
SOX6
|
SRY (sex determining region Y)-box 6
|
|
chr11_-_69343128
|
0.556
|
NM_005247
|
FGF3
|
fibroblast growth factor 3
|
|
chr3_+_45611180
|
0.555
|
NM_014240
|
LIMD1
|
LIM domains containing 1
|
|
chr1_+_22107137
|
0.553
|
|
RPL21
|
ribosomal protein L21
|
|
chr16_+_64958060
|
0.549
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr19_-_1543633
|
0.549
|
NM_003926
|
MBD3
|
methyl-CpG binding domain protein 3
|
|
chr16_+_64958025
|
0.548
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr3_-_50340598
|
0.547
|
|
TUSC2
|
tumor suppressor candidate 2
|
|
chr19_+_11510578
|
0.547
|
NM_001299
|
CNN1
|
calponin 1, basic, smooth muscle
|
|
chr17_+_43373887
|
0.546
|
NM_018129
|
PNPO
|
pyridoxamine 5'-phosphate oxidase
|
|
chr19_+_53664386
|
0.545
|
|
CYTH2
|
cytohesin 2
|
|
chr19_+_54872220
|
0.540
|
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr7_-_104696678
|
0.539
|
NM_182691
|
SRPK2
|
SRSF protein kinase 2
|
|
chr19_+_54872304
|
0.539
|
NM_001536
NM_198318
|
PRMT1
|
protein arginine methyltransferase 1
|
|
chr17_+_34114924
|
0.539
|
|
MLLT6
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 6
|
|
chr16_-_3008162
|
0.537
|
NM_021195
|
CLDN6
|
claudin 6
|
|
chr1_-_153560532
|
0.536
|
NM_001039517
|
C1orf104
|
chromosome 1 open reading frame 104
|
|
chr1_+_34997928
|
0.534
|
NM_153212
|
GJB4
|
gap junction protein, beta 4, 30.3kDa
|
|
chr4_-_40631397
|
0.533
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr12_+_47658502
|
0.532
|
NM_005430
|
WNT1
|
wingless-type MMTV integration site family, member 1
|
|
chr11_+_131285747
|
0.531
|
NM_001144058
NM_001144059
NM_016522
|
NTM
|
neurotrimin
|
|
chr1_+_201363458
|
0.530
|
NM_000674
NM_001048230
|
ADORA1
|
adenosine A1 receptor
|
|
chr19_+_12910428
|
0.529
|
|
CALR
|
calreticulin
|
|
chr16_-_88084387
|
0.528
|
NM_013275
|
ANKRD11
|
ankyrin repeat domain 11
|
|
chr3_+_45610357
|
0.525
|
|
LIMD1
|
LIM domains containing 1
|
|
chr22_+_17498320
|
0.524
|
NM_053006
|
TSSK2
|
testis-specific serine kinase 2
|
|
chr19_+_12910391
|
0.523
|
NM_004343
|
CALR
|
calreticulin
|
|
chr1_+_154855709
|
0.520
|
NM_021817
|
HAPLN2
|
hyaluronan and proteoglycan link protein 2
|
|
chr1_+_36394338
|
0.515
|
NM_018067
|
MAP7D1
|
MAP7 domain containing 1
|
|
chr12_+_91620749
|
0.514
|
NM_001037671
NM_001178097
|
C12orf74
|
chromosome 12 open reading frame 74
|
|
chr17_+_8154280
|
0.514
|
NM_173728
|
ARHGEF15
|
Rho guanine nucleotide exchange factor (GEF) 15
|
|
chr7_+_4781784
|
0.510
|
NM_014855
|
KIAA0415
|
KIAA0415
|
|
chr1_-_6468108
|
0.508
|
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr4_+_54660954
|
0.506
|
NM_133267
|
GSX2
|
GS homeobox 2
|
|
chr17_-_71017509
|
0.506
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr16_+_1773004
|
0.503
|
|
NUBP2
|
nucleotide binding protein 2 (MinD homolog, E. coli)
|
|
chr5_+_149845503
|
0.503
|
|
NDST1
|
N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1
|
|
chr16_+_22432344
|
0.502
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr16_-_65775326
|
0.500
|
NM_001040715
|
KIAA0895L
|
KIAA0895-like
|
|
chr12_+_111980044
|
0.498
|
NM_004416
|
DTX1
|
deltex homolog 1 (Drosophila)
|
|
chr4_+_87070449
|
0.497
|
NM_031305
|
ARHGAP24
|
Rho GTPase activating protein 24
|
|
chr7_+_150321771
|
0.495
|
NM_001160109
NM_001160110
NM_001160111
|
NOS3
|
nitric oxide synthase 3 (endothelial cell)
|
|
chr9_+_138341752
|
0.494
|
NM_001145638
NM_015597
|
GPSM1
|
G-protein signaling modulator 1
|
|
chr15_+_72397934
|
0.491
|
NM_182791
|
CCDC33
|
coiled-coil domain containing 33
|
|
chr3_+_14419066
|
0.490
|
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr12_+_48047428
|
0.488
|
|
SPATS2
|
spermatogenesis associated, serine-rich 2
|
|
chr5_+_140719773
|
0.484
|
NM_018923
NM_032096
|
PCDHGB2
|
protocadherin gamma subfamily B, 2
|
|
chr3_+_14419087
|
0.483
|
NM_001134367
NM_001134368
NM_003043
|
SLC6A6
|
solute carrier family 6 (neurotransmitter transporter, taurine), member 6
|
|
chr9_+_136358134
|
0.482
|
NM_002957
|
RXRA
|
retinoid X receptor, alpha
|
|
chr5_+_140767953
|
0.481
|
NM_018926
NM_032100
|
PCDHGB6
|
protocadherin gamma subfamily B, 6
|
|
chr9_+_94766379
|
0.480
|
|
FGD3
|
FYVE, RhoGEF and PH domain containing 3
|
|
chr16_+_64958086
|
0.479
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr17_+_72376022
|
0.478
|
NM_144677
|
MGAT5B
|
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B
|
|
chr10_+_70748581
|
0.478
|
NM_000188
|
HK1
|
hexokinase 1
|
|
chr3_+_195887903
|
0.476
|
NM_153690
|
FAM43A
|
family with sequence similarity 43, member A
|
|
chr9_+_132990760
|
0.475
|
NM_005085
|
NUP214
|
nucleoporin 214kDa
|
|
chr9_+_124590970
|
0.471
|
NM_001001923
|
OR5C1
|
olfactory receptor, family 5, subfamily C, member 1
|
|
chr5_+_140573692
|
0.470
|
NM_018933
|
PCDHB13
|
protocadherin beta 13
|
|
chr6_-_42527760
|
0.470
|
NM_033502
|
TRERF1
|
transcriptional regulating factor 1
|
|
chr13_+_48923688
|
0.470
|
NM_001160308
|
SETDB2
|
SET domain, bifurcated 2
|
|
chr5_+_140777454
|
0.469
|
NM_018927
NM_032101
|
PCDHGB7
|
protocadherin gamma subfamily B, 7
|
|
chr15_+_80434362
|
0.468
|
|
UBE2Q2P2
|
ubiquitin-conjugating enzyme E2Q family member 2 pseudogene 2
|
|
chr6_+_50789215
|
0.467
|
NM_172238
|
TFAP2D
|
transcription factor AP-2 delta (activating enhancer binding protein 2 delta)
|
|
chr2_-_100088760
|
0.466
|
|
AFF3
|
AF4/FMR2 family, member 3
|
|
chr19_+_12810258
|
0.463
|
NM_014975
|
MAST1
|
microtubule associated serine/threonine kinase 1
|
|
chr17_+_39778191
|
0.463
|
|
GRN
|
granulin
|
|
chr17_-_1478293
|
0.461
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr1_+_12757570
|
0.461
|
NM_001080830
|
PRAMEF12
|
PRAME family member 12
|
|
chr1_+_149299490
|
0.460
|
|
MLLT11
|
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11
|
|
chr16_+_57091546
|
0.459
|
|
NDRG4
|
NDRG family member 4
|
|
chr6_+_32515596
|
0.456
|
NM_019111
|
HLA-DRA
|
major histocompatibility complex, class II, DR alpha
|
|
chr22_+_48633532
|
0.455
|
|
ZBED4
|
zinc finger, BED-type containing 4
|
|
chr19_+_50196526
|
0.455
|
NM_006509
|
RELB
|
v-rel reticuloendotheliosis viral oncogene homolog B
|
|
chr19_-_16544188
|
0.455
|
NM_024881
|
SLC35E1
|
solute carrier family 35, member E1
|
|
chr17_+_19930926
|
0.450
|
NM_001033553
NM_152904
|
SPECC1
|
sperm antigen with calponin homology and coiled-coil domains 1
|